Example of custom scan_info¶
This examples show how to describe metadata in scan_info for few specific
scans.
The main use case is to: - Allow to display scatter as a solid regular 2D image. - Display the entire region of the scan before the end
Displaying scatter data as a scatter of points is not a problem.
With this extra metadata, the user can decide to use a solid or point based rendering.
Setup¶
This scans have to be created usually per beamline in a very specific way.
For this examples we will use a function to simulate them.
import gevent
import numpy
from bliss.scanning.group import Sequence
from bliss.scanning.chain import AcquisitionChannel
def simulate_scan(scan_info, data):
"""
Simulate a scan.
It uses a sequence to emit custom channels.
Arguments:
scan_info: Dictionary contanining metadata to store with the scan
data: Dictionary containing numpy array per channel name
"""
step = 2
seq = Sequence(scan_info=scan_info)
for channel_name, array in data.items():
seq.add_custom_channel(AcquisitionChannel(channel_name, float, ()))
with seq.sequence_context() as scan_seq:
array = list(data.values())[0]
for i in range(0, len(array) - 1, step):
selection = slice(i, i + step)
for channel_name, array in data.items():
seq.custom_channels[channel_name].emit(array[selection])
gevent.sleep(0.1)
Regular scatter¶
This example create a regular scatter.
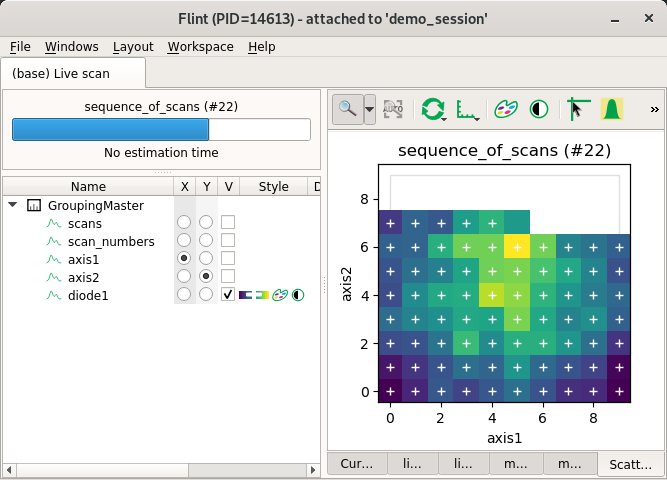
from bliss.scanning.scan_info import ScanInfo
import numpy
scan_info = ScanInfo()
# Specify the same group for all this channels (axis or values)
scan_info.set_channel_meta("axis1",
# The group have to be the same for all this channels
group="foo",
# This is the fast axis
axis_id=0,
# In forth direction only
axis_kind="forth",
# The grid have to be specified
start=0, stop=9, axis_points=10,
# Optionally the full number of points can be specified
points=100)
scan_info.set_channel_meta("axis2", group="foo", axis_id=1,
axis_kind="forth",
start=0, stop=9,
axis_points=10, points=100)
scan_info.set_channel_meta("diode1", group="foo")
# Request a specific scatter to be displayed
scan_info.add_scatter_plot(x="axis1", y="axis2", value="diode1")
# Generate the data
slow, fast = numpy.mgrid[0:10, 0:10]
dist = (((slow - 4.5) ** 2 + (fast - 4.5) ** 2) ** 0.5)
dist = (dist.max() - dist) * 10
data = {
"axis1": fast.flatten(),
"axis2": slow.flatten(),
"diode1": numpy.random.poisson(dist.flatten())
}
SCAN_DISPLAY.auto = True
simulate_scan(scan_info, data)
Regular 3D scatter¶
3D of more dimensions are supported.
Flint only can display it using the 2 fastest axes.
Other axes have to be stepper and are considered as an ndim index for the displayed frame.
displayed.
For now only Flint will only display the last frame.
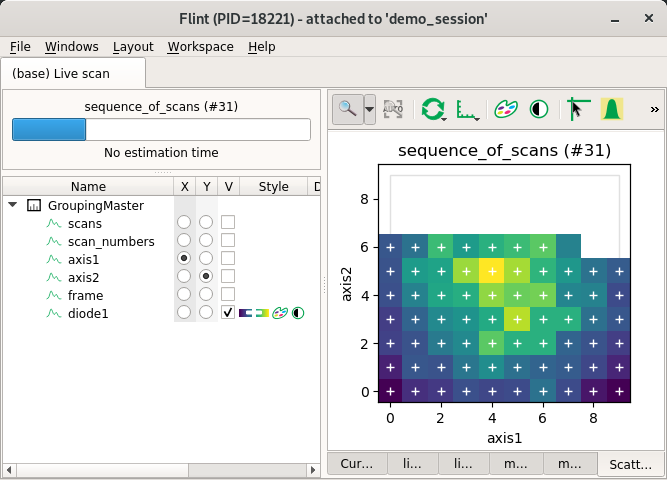
from bliss.scanning.scan_info import ScanInfo
import numpy
scan_info = ScanInfo()
scan_info.set_channel_meta("axis1", group="foo", axis_id=0,
axis_kind="forth",
start=0, stop=9, axis_points=10,
points=1000)
scan_info.set_channel_meta("axis2", group="foo", axis_id=1,
axis_kind="forth",
start=0, stop=9, axis_points=10,
points=1000)
# The slowest axes have to be describe as stepper
scan_info.set_channel_meta("frame", group="foo", axis_id=2,
axis_kind="step",
start=0, stop=9, axis_points=10,
points=1000)
scan_info.set_channel_meta("diode1", group="foo")
# Request a specific scatter to be displayed
scan_info.add_scatter_plot(x="axis1", y="axis2", value="diode1")
# Generate the data
frame, slow, fast = numpy.mgrid[0:10, 0:10, 0:10]
dist = (((slow - 4.5) ** 2 + (fast - 4.5) ** 2) ** 0.5)
dist = (dist.max() - dist) * 10
data = {
"axis1": fast.flatten(),
"axis2": slow.flatten(),
"frame": frame.flatten(),
"diode1": numpy.random.poisson(dist.flatten())
}
SCAN_DISPLAY.auto = True
simulate_scan(scan_info, data)
Theorical and encoder position together in scatter¶
The same axis can be describe many time using many channels.
That’s what we simulate here. axis1 and axis2 are also provided as
axis1-enc, axis2-enc.
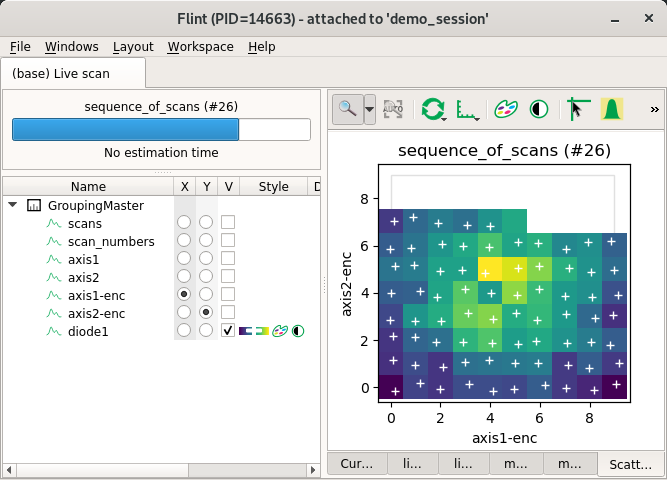
from bliss.scanning.scan_info import ScanInfo
import numpy
scan_info = ScanInfo()
scan_info.set_channel_meta("axis1", group="foo", axis_id=0, axis_kind="forth",
start=0, stop=9, axis_points=10, points=100)
scan_info.set_channel_meta("axis2", group="foo", axis_id=1, axis_kind="forth",
start=0, stop=9, axis_points=10, points=100)
# Index are reused for the encoded
scan_info.set_channel_meta("axis1-enc", group="foo", axis_id=0, axis_kind="forth",
start=0, stop=9, axis_points=10, points=100)
scan_info.set_channel_meta("axis2-enc", group="foo", axis_id=1, axis_kind="forth",
start=0, stop=9, axis_points=10, points=100)
scan_info.set_channel_meta("diode1", group="foo")
# Request a specific scatter to be displayed
scan_info.add_scatter_plot(x="axis1", y="axis2", value="diode1")
# Generate the data
slow, fast = numpy.mgrid[0:10, 0:10]
dist = (((slow - 4.5) ** 2 + (fast - 4.5) ** 2) ** 0.5)
dist = (dist.max() - dist) * 10
data = {
"axis1": fast.flatten(),
"axis2": slow.flatten(),
"axis1-enc": fast.flatten() + (numpy.random.rand(100) * 0.4 - 0.2),
"axis2-enc": slow.flatten() + (numpy.random.rand(100) * 0.4 - 0.2),
"diode1": numpy.random.poisson(dist.flatten())
}
SCAN_DISPLAY.auto = True
simulate_scan(scan_info, data)
Back and forth axis scatter¶
For now Flint is not able to display it with a solid rendering. It could be displayed as a regular grid.
For now you can use metadata for non-regular scatters.
Non-regular scatters¶
Flint can display non regular scatters with solid rendering using 2D histogram.
This create a regular 2D mesh, and each cells can contain more than one value.
The cell (the pixel) value is computed using a reduction function (for now it
uses mean function).
Hint have to be provided to describe the size of the histogram grid.
axis-points can not be specified because the axis is not regular.
But axis-points-hint have to be used (with start and stop) and will be
used to know the amount of pixels to used per axis.
start and stop have maybe no meaning here, so you can use min and max.
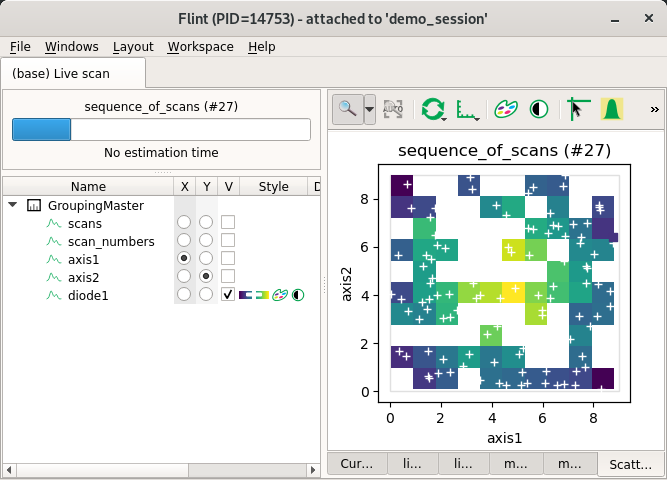
from bliss.scanning.scan_info import ScanInfo
import numpy
scan_info = ScanInfo()
# There is no hint for axis kind, and axis npoints is a hint
scan_info.set_channel_meta("axis1", group="foo", axis_id=0,
min=0, max=9, axis_points_hint=10, points=500)
scan_info.set_channel_meta("axis2", group="foo", axis_id=1,
min=0, max=9, axis_points_hint=10, points=500)
scan_info.set_channel_meta("diode1", group="foo")
# Request a specific scatter to be displayed
scan_info.add_scatter_plot(x="axis1", y="axis2", value="diode1")
# Generate the data
axis1 = numpy.random.rand(500) * 9
axis2 = numpy.random.rand(500) * 9
frame = numpy.mgrid[0:5, 0:100][0]
dist = (((axis1 - 4.5) ** 2 + (axis2 - 4.5) ** 2) ** 0.5)
dist = (dist.max() - dist) * 10
data = {
"axis1": axis1,
"axis2": axis2,
"diode1": numpy.random.poisson(dist)
}
SCAN_DISPLAY.auto = True
simulate_scan(scan_info, data)
Non-regular 3D scatters¶
This also can work for extra dimensions with steppers.
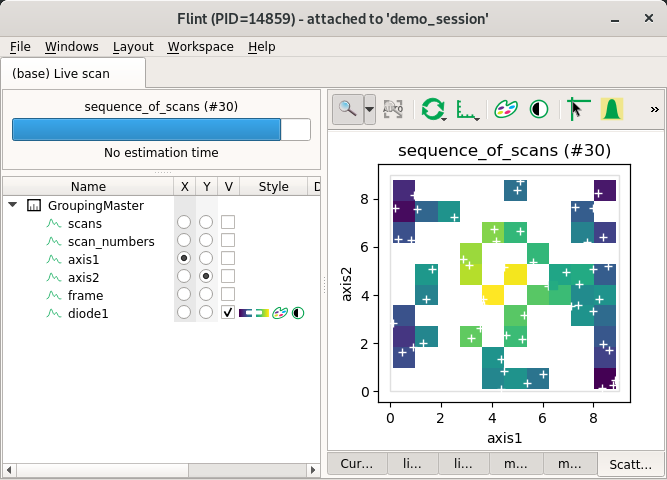
from bliss.scanning.scan_info import ScanInfo
import numpy
scan_info = ScanInfo()
# There is no hint for axis kind, and axis npoints is a hint
scan_info.set_channel_meta("axis1", group="foo", axis_id=0,
min=0, max=9, axis_points_hint=10, points=500)
scan_info.set_channel_meta("axis2", group="foo", axis_id=1,
min=0, max=9, axis_points_hint=10, points=500)
scan_info.set_channel_meta("frame", group="foo", axis_id=2,
axis_kind="step",
start=0, stop=4, axis_points=5,
points=500)
scan_info.set_channel_meta("diode1", group="foo")
# Request a specific scatter to be displayed
scan_info.add_scatter_plot(x="axis1", y="axis2", value="diode1")
# Generate the data
axis1 = numpy.random.rand(500) * 9
axis2 = numpy.random.rand(500) * 9
frame = numpy.mgrid[0:5, 0:100][0]
dist = (((axis1 - 4.5) ** 2 + (axis2 - 4.5) ** 2) ** 0.5)
dist = (dist.max() - dist) * 10
data = {
"axis1": axis1,
"axis2": axis2,
"frame": frame.flatten(),
"diode1": numpy.random.poisson(dist)
}
SCAN_DISPLAY.auto = True
simulate_scan(scan_info, data)