Defaults BLISS scans¶
BLISS step-by-step scan functions¶
BLISS provides functions to perform step-by-step scans. The acquisition
chain for those scans is built using the DefaultChain class.
Parameters (common to all scans)¶
Counters parameters¶
counter_args (counter-providing objects): each parameter provides
counters to be integrated in the scan. if no counter parameters are
provided, use the active measurement group.
Optional parameters¶
Common keyword arguments that can be used in all scans:
name (str): scan name in data nodes tree and directories [default: ‘scan’]title (str): scan title [default: ‘a2scan… ‘] save (bool): save scan data to file [default: True]save_images (bool or None): save image files [default: None, means it follows the save argument]sleep_time (float): sleep time between 2 points [default: None]stab_time (float): stabilization time after a move, before counting [default: None]run (bool): ifTrue(default), run the scan.Falsemeans to just create scan object and acquisition chain.return_scan (bool): [default: True by default]
sleep time
sleep_time sleeping time is done between points of the scan, not before
the first one.
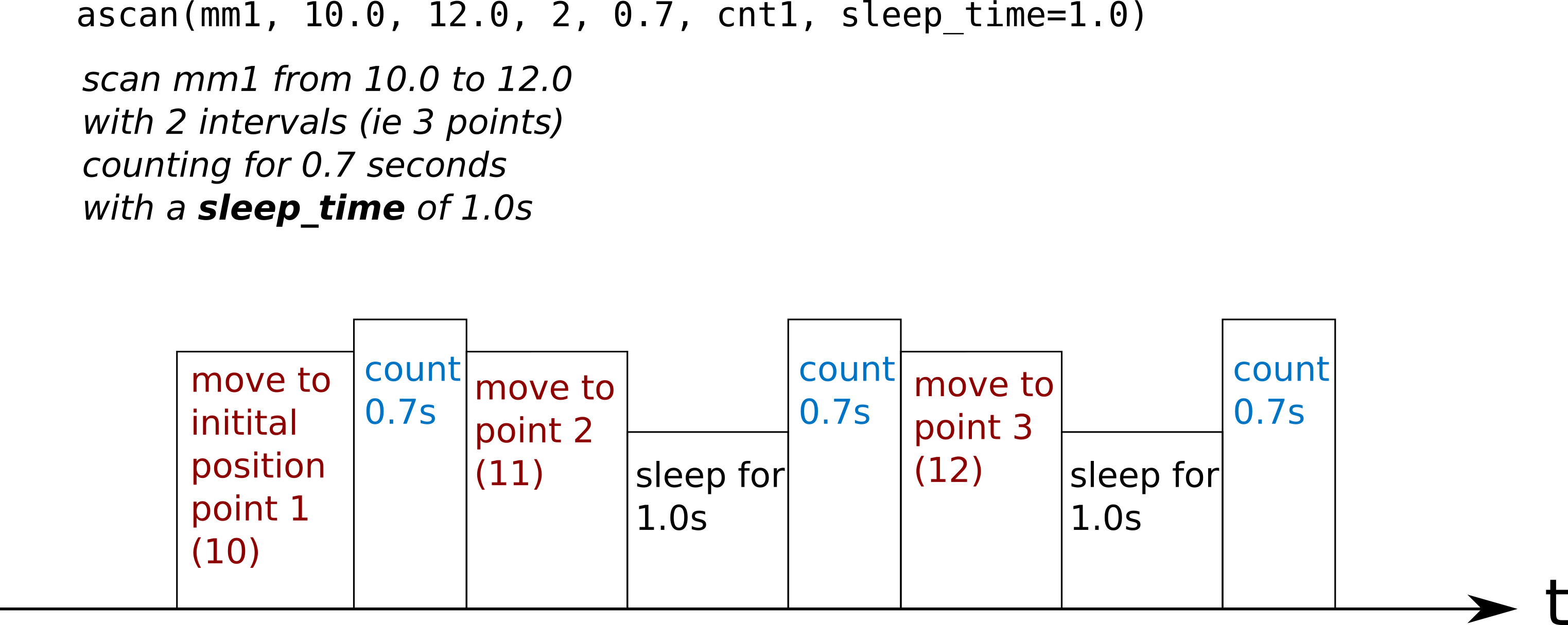
stabilization time
stab_time sleeping time is done before each count even before the
first of the scan.
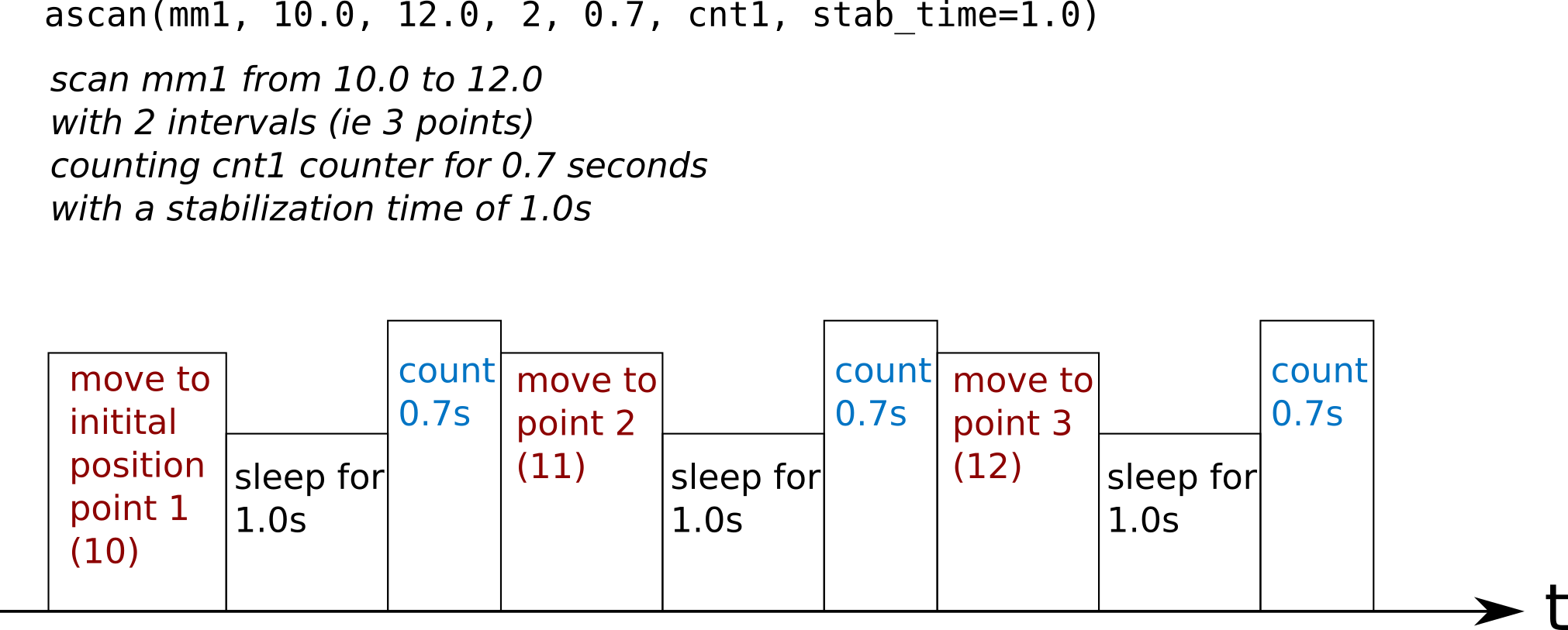
Note
As stab_time concerns movement, it does not apply in timescan and loopscan.
Scan example¶
ascan(<mot>, <start>, <stop>, <intervals>, <acq_time>, <title>)
ascan( sy, 1, 2, 4, 0.5, title="Jadarite_LiNaSiB3O7(OH)")
This command performs a scan of five 500ms-counts of current measurement group
counters at <sy> motor positions: 1, 1.25, 1.5, 1.75 and 2.
ascan dscan¶
ascan(motor, start, stop, intervals, count_time, *counter_args, **kwargs)
Absolute scan of one motor, as specified by <motor>. The motor starts at
the position given by <start> and ends at the position given by <stop>.
The step size is: (<stop>-<start>)/<intervals>
The number of points will be <intervals> + 1.
Count time is given by <count_time> (seconds).
At the end of the scan, the motor will stay at stopping position
(<stop> position in case of success).
Idem for dscan but using relative positions:
dscan(motor, rel_start, rel_stop, intervals, count_time, *counter_args, **kwargs)
Scans one motor, as specified by <motor>. If the motor is at position X
before the scan begins, the scan will run from X+rel_start to X+rel_stop.
The step size is: (<rel_stop>-<rel_start>)/<intervals>. The number of points
will be <intervals>+1. Count time is given by <count_time> (in seconds).
Note
At the end of a dscan (even in case of error or scan abortion, on a ctrl-c
for example) the motor will return to its initial position.
Note
A dscan will be rewriten in hdf5 file as an ascan: relative positions
will be replaced by absolute ones.
a2scan¶
a2scan( motor1, start1, stop1,
motor2, start2, stop2,
intervals, count_time, *counter_args, **kwargs)
Absolute 2 motors scan.
Scans two motors, as specified by <motor1> and <motor2>. The
motors start at the positions given by <start1> and <start2> and
end at the positions given by <stop1> and <stop2>. The step size
for each motor is given by (<stopN>-<startN>)/<intervals>. The
number of points will be <intervals>+1. Count time is given by
<count_time> (in seconds).
d2scan¶
Relative 2 motors scan.
Scans two motors, as specified by <motor1> and <motor2>. Each motor moves
the same number of points. If a motor is at position X
before the scan begins, the scan will run from X+<start> to X+<end>.
The step size of a motor is (<stopN>-<startN>)/<intervals>. The number
of points will be <intervals>+1. Count time is given by <count_time>
(in seconds).
At the end of the scan (even in case of error) the motors will return to their initial positions.
a3scan a4scan a5scan¶
Similary to a2scan, aNscan functions are provided fo N in {3,4,5}.
example for 9 intervals:
DEMO [2]: a5scan(m1,1,2, m2,3,4, m3,5,6, m4,7,8, m5,8,9, 9, 0.1)
Scan 2 Fri Oct 26 16:07:08 2018 /tmp/scans/demo/
a5scan m1 1 2 m2 3 4 m3 5 6 m4 7 8 m5 8 9 10 0.1
# dt[s] m1 m2 m3 m4 m5 simct1
0 0 1 3 5 7 8 0.038648
1 0.432173 1.11 3.11 5.11 7.11 8.11 0.022345
2 0.850866 1.22 3.22 5.22 7.22 8.22 0.119345
3 1.25996 1.33 3.33 5.33 7.33 8.33 1.06995
4 1.74734 1.44 3.44 5.44 7.44 8.44 3.45354
5 2.16594 1.56 3.56 5.56 7.56 8.56 3.47793
6 2.57817 1.67 3.67 5.67 7.67 8.67 1.01595
7 2.98574 1.78 3.78 5.78 7.78 8.78 0.128783
8 3.4303 1.89 3.89 5.89 7.89 8.89 0.073870
9 3.84454 2 4 6 8 9 0.019919
Took 0:00:05.226827
Out [2]: Scan(name=a5scan_2, run_number=2, path=/tmp/scans/demo/)
aNscan dNscan¶
In case a scan for more than 5 motors is needed, anscan and dnscan functions
can be used with a slightly different list of parameters:
anscan([(<mot1>, <start1>, <stop1>),...,(<motN>, <startN>, <stopN>)], <intervals>, <counting_time>, counter1, ... counterN )
(<mot>, <start>, <stop>) can be repeated as much as needed.
idem for dnscan with relative start and stop positions:
dnscan([(<mot1>, <start1>, <stop1>),...,(<motN>, <startN>, <stopN>)], <intervals>, <counting_time>, counter1, ... counterN )
amesh¶
amesh( motor1, start1, stop1, intervals1,
motor2, start2, stop2, intervals2,
count_time, *counter_args, **kwargs)
Mesh scan.
The amesh scan traces out a grid using motor <motor1> and motor
<motor2>. The first motor scans from position <start1> to <end1> using the
specified number of intervals + 1 as points number. The second motor similarly
scans from <start2> to <end2>. Each point is counted for for <count_time>
seconds (or monitor counts).
The scan of <motor1> is done at each point scanned by <motor2>. That is, the
first motor scan is nested within the second motor scan. (<motor1> is the
“fast” axis, <motor2> the “slow” axis)
Special parameter:
backnforth: if True, do back and forth on the first motor.
a3mesh¶
The a3mesh scan traces out a grid using motor1, motor2 and motor3. Each
motor uses its own specified start, stop and intervals. Each point is
counted for time seconds (or monitor counts).
The scan of motor1 is done at each point scanned by motor2. The scan of
motor1+motor2 is done at each point scanned by motor3. That is, the first
motor scan is nested within the second motor scan which is nested within the
third motor.
Special parameter:
backnforth: if True do back and forth on the first 2 motors
other mesh¶
dmesh: Relative amesh.d3mesh: Relative mesh with 3 motors
anmesh¶
Mesh scan with n-motors
This scan traces out a grid using all the motors. Each motor uses its own
specified start, stop and intervals. Each point is counted for time
seconds (or monitor counts).
The first motor is the fastest, and the last motor the slowest. Each motor is nested with the next one.
Special parameters:
motor_tuple_list: List of tuple(motor, start, stop, interval). The first motor is the fastest.backnforth: If True do back and forth for all the motors except the slowest one
lineup¶
Relative scan.
lineup(motor, start, stop, intervals, count_time, counter, **kwargs)
lineup performs a dscan and then goes to the maximum value of the counter.
It only accepts one single counter as argument
timescan¶
Scan without movement.
timescan(count_time, *counter_args, **kwargs)
Performs <npoints> counts for <count_time>. If <npoints> is 0, it
counts forever.
Special parameters:
<output_mode> (str): valid are ‘tail’ (append each line to output) or ‘monitor’ (refresh output in single line) [default: ‘tail’]<npoints> (int): number of points [default: 0, meaning infinite number of points]
loopscan¶
loopscan(npoints, count_time, *counter_args, **kwargs)
Similar to timescan but <npoints> is mandatory.
ct¶
ct(count_time, *counter_args, **kwargs)
Counts for a specified time.
Warning
ct serves for beamline snapshots. It does neither collect any metadata
nor offers the possibilty to save the results. Use sct instead.
sct¶
sct(count_time, *counter_args, **kwargs)
Similar to ct.
Counts for a specified time and saves the results like any other scan.
pointscan¶
Performs a scan over many positions given as a list.
pointscan(motor, positions_list, count_time, *counter_args, **kwargs)
Scans one motor, as specified by <motor>. The motor starts at the
position given by the first value in <positions_list> and ends at the
position given by last value <positions_list>. Count time is given by
<count_time> (in seconds).
Special parameter:
<positions_list>: List of positions to scan for<motor>motor.
pointscan is based on lookupscan, reducing it to only one motor.
lookupscan¶
Previous multi-motors scans (aNscan, dNscan) are based on the generic
lookupscan. It can take a variable number of motors associated to a
list of positions to use for the scan.
usage:
lookupscan([(<mot_1>, <positions_list_1>),...,(<mot_N>, <positions_list_N>)], counting_time, <counter>*, **kwargs)
example:
import numpy as np
lookupscan([(m0, np.arange(0, 2, 0.5)),(m1, np.linspace(1, 3, 4))], 0.1, diode2)
Scans behaviour¶
- create acquisition device
prepare()start()trigger()called<nbpoints>times.stop()
| Scan | nbpoints | start/stop type |
|---|---|---|
| def. | N | list |
| timescan | 0 | [] |
| loopscan | N | [] |
| pointscan | N | float |
| ct | 1 | [] |
Default chain¶
All standard scans (step scans) are built the same way using the
DefaultAcquisitionChain object accessible via the global variable
DEFAULT_CHAIN, if you are in a session.
This object builds the acquisition chain with the default top masters and the acquisition objects with their default acquisition parameters.
The default top masters are the SoftwareTimer (ct, loopscan, timescan)
or one of the motor masters (VariableStepTriggerMaster, MeshStepTriggerMaster)
for the default scans working with axes (ascan, amesh, pointscan, lookupscan).
The default acquisition parameters for an acquisition object are defined in the
associated controller class (see get_default_chain_parameters).
These defaults can be customized via the configuration (.yml) and activated
using the set_settings method of the DEFAULT_CHAIN.
Be aware that it will affect all standard scans permanently.
Below an example of a YAML configuration file with two basler cameras customized to receive an hardware trigger provided by a p201 counting card:
- name: default_acq_chain
plugin: default
chain_config:
- device: $basler_1
acquisition_settings:
acq_trigger_mode: EXTERNAL_TRIGGER_MULTI
master: $p201_0
- device: $basler_2
acquisition_settings:
acq_trigger_mode: EXTERNAL_TRIGGER_MULTI
master: $p201_0
To activate this settings for all standard scan of your session do as follows in the session setup file :
DEFAULT_CHAIN.set_settings(default_acq_chain['chain_config'])
Two types of customization are possible:
- Modify the default
acquisition_settings(i.e. acquisition parameters) of an acquisition object associated to adevice.
- device: $basler_1
acquisition_settings:
acq_trigger_mode: EXTERNAL_TRIGGER_MULTI
...
- Add a
masteron top of adevice.
- device: $basler_1
...
master: $p201_0
The master device can also be customized by adding a device key for this master.
- device: $p201_0
acquisition_settings:
acq_mode: ExtTrigMulti
Note
Acquisition parameters are all the parameters that define the
number of triggers and points, trigger type, exposure time and so on.
Other detector parameters should not be part of the DefaultAcquisitionChain
configuration.
For example Image configuration (binning, flip, rotation) or Saving parameters of a Lima device, should be excluded from this configuration and set before any scan.
ChainPreset can also be added to
the DEFAULT_CHAIN. Usually this is also done in the session setup
like this:
DEFAULT_CHAIN.add_preset(my_preset)
To go further…¶
Steps scans¶
Most unusual step scans can be defined using one of the existing standard scans.
n-regions scan example¶
In this example you want to define a scan with several regions. The regions have to be defined as a list of tuples like: [(start1,stop1,npoints1),(start2,stop2,npoints2),…]
import numpy
from bliss.common.scans import pointscan
def n_region_scan(motor, regions, count_time, *counter_args, **kwargs):
positions = list()
for start,stop,npoints in regions:
positions.extend(numpy.linspace(start,stop,npoints))
# change to new defined scan
kwargs.setdefault('type', f'{len(regions)}_region_scan')
# Build a **meaning** title
kwargs.setdefault('title',f'{kwargs.get("type")} on {motor.name}')
return pointscan(motor,positions,count_time,*counter_args,**kwargs)
Execute :
DEMO [1]: s = n_region_scan(roby,[(0,2,3),(10,15,11)],0.1,diode,save=False)
Scan 9 Tue Apr 02 14:58:33 2019 <no saving> demo user = seb
2_region_scan on roby
# dt[s] roby diode
0 0 0 23.8889
1 0.232985 1 23.5556
2 0.46471 2 -2.11111
3 0.823396 10 16.5556
4 1.01696 10.5 -8.88889
5 1.20862 11 -17.3333
6 1.39964 11.5 20.6667
7 1.59078 12 0.444444
8 1.78076 12.5 17.7778
9 1.97064 13 -0.111111
10 2.16226 13.5 26
11 2.35261 14 28.2222
12 2.54606 14.5 -1.55556
13 2.7383 15 59.2222
Took 0:00:03.366149
ascans, which take step size rather than the number of intervals¶
import numpy
from bliss.common.scans import ascan
def step_scan(motor, start, stop, step_size, count_time, *counter_args, **kwargs):
intervals = int(numpy.ceil(abs(start-stop)/step_size)) - 1
return ascan(motor, start, stop, intervals, count_time, *counter_args, **kwargs)
Execute :
TEST_SESSION [42]: s = step_scan(roby, 0, 1, 0.2, 0.1, diode)
Scan 17 Tue Apr 02 16:04:31 2019 /tmp/..../data.h5 test_session user = seb
ascan roby 0 1 5 0.1
# dt[s] roby diode
0 0 0 -9.33333
1 0.193154 0.25 -38.5556
2 0.385237 0.5 -6.55556
3 0.575978 0.75 -13.5556
4 0.765097 1 -9.55556
Took 0:00:01.229621
Using ‘presets’ to customize a scan¶
In this example, the scan will pump a certain amount of liquid using a syringe before each point. To do this we will use ChainPreset.
from bliss.scanning.chain import ChainPreset,ChainIterationPreset
from bliss.common.scans import ascan
class Syringe:
def __init__(self, available_liquid):
self._available_liquid = available_liquid
def pump(self,amount):
if self._available_liquid < amount:
raise RuntimeError("No more liquid to pump")
self._available_liquid -= amount
my_syringe = Syringe(10) # liquid volume == 10
def syringe_ascan(syringe, liquid_amount,
motor, start, stop, intervals, count_time, *counter_args, **kwargs):
class Preset(ChainPreset):
class Point(ChainIterationPreset):
def prepare(self):
syringe.pump(liquid_amount)
def get_iterator(self,acq_chain):
while True:
yield Preset.Point()
kwargs.setdefault('run',False)
s = ascan(motor, start, stop, intervals, count_time, *counter_args, **kwargs)
preset = Preset()
s.acq_chain.add_preset(preset)
s.run()
return s
Execute :
TEST_SESSION [16]: syringe_ascan(my_syringe, 1, roby, 0, 1, 15, 0.1, diode)
Scan 22 Tue Apr 02 16:37:24 2019 /tmp/..../data.h5 test_session user = seb
ascan roby 0 1 15 0.1
# dt[s] roby diode
0 0 0 12.6667
1 0.19153 0.0714 -12.2222
2 0.381468 0.1429 9
3 0.570563 0.2143 11.6667
4 0.761761 0.2857 -12.1111
5 0.953436 0.3571 10.8889
6 1.14317 0.4286 -1.44444
7 1.33038 0.5 -21.3333
8 1.51747 0.5714 51.3333
9 1.7079 0.6429 9.77778
!!! === RuntimeError: No more liquid to pump === !!!
Took 0:00:02.008699
!!! === RuntimeError: No more liquid to pump === !!!
In this example, before each point preparation the syringe will pump one unit of a volume and raises an error when the syringe is empty.
Note
Any exception in Preset method stop the scan.
More complex scans¶
For more complex scans, you may need to use a lower level api. see: Scan engine.