Dataset metadata
Under the ESRF data policy, scans are grouped together in a dataset.
In BLISS, datasets are represented as Dataset objects that can be
accessed via SCAN_SAVING.dataset. These objects map to a group of
scans in REDIS and also collect the associated ICAT metadata.
Note
This only exists for the ESRF data policy
Warning
Dataset metadata is not saved in the HDF5 files. It is sent to the ESRF data policy services and is meant for searching datasets in the data portal
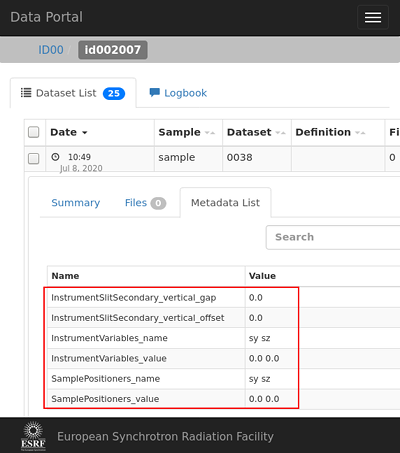
Configuration¶
Dataset metadata is collected from all controllers that implement the
HasMetadataForDataset protocol (ie: they inherit from HasMetadataForDataset
class).
Optionally, dataset metadata can be configured in the beamline configuration.
Available metadata¶
Dataset metadata is stored in the ICAT database. This database contains a fixed number offields.
These fields can be explored through the ICAT definition object, which can be accessed through one of:
demo.icat_metadata.definitions(“demo” is the name of the Bliss session)SCAN_SAVING.dataset.definitionsSCAN_SAVING.collection.definitionsSCAN_SAVING.proposal.definitions
For shell usage, there is also SCAN_SAVING.dataset.existing that can be used to
view and modify the current dataset metadata
DEMO [2]: SCAN_SAVING.dataset.existing
Out [2]: Namespace contains:
.InstrumentVariables_name ['sy', 'sz']
.InstrumentVariables_value [0.0, 0.0]
.InstrumentSlitSecondary_vertical_gap 0.0
.InstrumentSlitSecondary_vertical_offset 0.0
.SamplePositioners_name ['sy', 'sz']
.SamplePositioners_value [0.0, 0.0]
.FLUO_i0 17.1
Add metadata¶
Manually¶
Use SCAN_SAVING.dataset.metadata to add custom metadata (has auto completion in the shell)
DEMO [1]: SCAN_SAVING.dataset.metadata.instrument.detector01.name = "eiger1"
or equivalently, if you know the ICAT field name:
DEMO [2]: SCAN_SAVING.dataset["InstrumentDetector01_name"] = "eiger1"
or:
DEMO [3]: SCAN_SAVING.all.InstrumentDetector01_name = "eiger1"
Existing metadata can also be modified like this (has auto completion in the shell):
DEMO [4]: SCAN_SAVING.dataset.existing.InstrumentDetector01_name = "eiger1"
Expected metadata can also be modified like this (has auto completion in the shell):
DEMO [5]: SCAN_SAVING.dataset.expected.InstrumentDetector01_name = "eiger1"
Techniques¶
To add the techniques used to collect a dataset:
DEMO [1]: SCAN_SAVING.dataset.add_techniques("fluo", "xrpd")
DEMO [2]: SCAN_SAVING.dataset.techniques
Out [2]: {'FLUO', 'XRPD'}
Some techiques also have additional metadata fields:
DEMO [3]: SCAN_SAVING.dataset.definitions.FLUO.i0 = 17.1
Controller¶
Controllers that provide dataset metadata should implement the HasMetadataForDataset
protocol. In particular, the method dataset_metadata, which should return a directory
of metadata. To know what the available keys are you can do this (for example for
bliss.controllers.motors.slits.Slits):
DEMO [2]: demo.icat_metadata.definitions.instrument.primary_slit
Out [2]: Namespace contains:
.name = IcatField(name='name', field_name='InstrumentSlitPrimary_name', parent='instrument.primary_slit', nxtype='NX_CHAR', description=None, units=None)
.vertical_gap = IcatField(name='vertical_gap', field_name='InstrumentSlitPrimary_vertical_gap', parent='instrument.primary_slit', nxtype='NX_CHAR', description=None, units=None)
.vertical_offset = IcatField(name='vertical_offset', field_name='InstrumentSlitPrimary_vertical_offset', parent='instrument.primary_slit', nxtype='NX_CHAR', description=None, units=None)
.horizontal_gap = IcatField(name='horizontal_gap', field_name='InstrumentSlitPrimary_horizontal_gap', parent='instrument.primary_slit', nxtype='NX_CHAR', description=None, units=None)
.horizontal_offset = IcatField(name='horizontal_offset', field_name='InstrumentSlitPrimary_horizontal_offset', parent='instrument.primary_slit', nxtype='NX_CHAR', description=None, units=None)
.blade_up = IcatField(name='blade_up', field_name='InstrumentSlitPrimary_blade_up', parent='instrument.primary_slit', nxtype='NX_CHAR', description=None, units=None)
.blade_down = IcatField(name='blade_down', field_name='InstrumentSlitPrimary_blade_down', parent='instrument.primary_slit', nxtype='NX_CHAR', description=None, units=None)
.blade_front = IcatField(name='blade_front', field_name='InstrumentSlitPrimary_blade_front', parent='instrument.primary_slit', nxtype='NX_CHAR', description=None, units=None)
.blade_back = IcatField(name='blade_back', field_name='InstrumentSlitPrimary_blade_back', parent='instrument.primary_slit', nxtype='NX_CHAR', description=None, units=None)
So dataset_metadata should return a dictionary, which has the keys name,
vertical_gap, vertical_offset, etc.
Dataset/sample/collection¶
The methods newdataset, newcollection and newsample will define the values of
some dataset metadata fields related to the dataset, collection and sample.
Dataset specific¶
The sample name and description can be defined for each dataset:
DEMO [1]: newdataset("dataset name", sample_name="my sample name", sample_description="my sample description")
This is equivalent to setting these two metadata fields:
DEMO [2]: SCAN_SAVING.dataset["Sample_name"] = "my sample name"
DEMO [3]: SCAN_SAVING.dataset["Sample_description"] = "my sample description"
Collection defaults¶
The sample name and description can be defined for each collection/sample:
DEMO [1]: newcollection("collection_name", sample_name="sample name", sample_description="my sample description")
or:
DEMO [2]: newsample("sample name", sample_description="my sample description")
All datasets belonging to this collection/sample will inherit this metadata. See further for details. This is equivalent to setting these two metadata fields for each dataset under this collection/sample:
DEMO [2]: SCAN_SAVING.dataset["Sample_name"] = "my sample name"
DEMO [3]: SCAN_SAVING.dataset["Sample_description"] = "my sample description"
Metadata inheritance¶
Metadata fields which are set on SCAN_SAVING.collection or SCAN_SAVING.proposal
will be used as defaults for SCAN_SAVING.dataset. This is for example how the
sample name is managed:
DEMO [10]: SCAN_SAVING.collection["Sample_name"] = "my sample"
DEMO [11]: SCAN_SAVING.dataset.existing
Out [11]: Namespace contains:
.Sample_name 'my sample'
DEMO [12]: SCAN_SAVING.dataset["Sample_name"] = "other name"
DEMO [13]: SCAN_SAVING.dataset.existing
Out [13]: Namespace contains:
.Sample_name 'other name'
DEMO [14]: newdataset()
DEMO [16]: SCAN_SAVING.dataset.existing
Out [17]: Namespace contains:
.Sample_name 'my sample'
So, the value of the Sample_name metadata field of a dataset is the value set on the collection,
when not specified explicitly for the dataset itself. This logic applies to all metadata fields.
Custom datasets¶
In this example, an isolated dataset for a dedicated experimental procedure is created
by using a custom ScanSaving object. This may make sense, when adding technique
related metadata (FLUO definition in this case).
from bliss.common.scans import loopscan
from bliss.setup_globals import diode1
from bliss.setup_globals import diode2
from bliss.setup_globals import mca1
from bliss.scanning.scan_saving import ScanSaving
from bliss import current_session
from bliss.scanning.scan import Scan
from pprint import pprint
def demo_with_technique():
"""a demo procedure using a custom scan saving"""
scan_saving = ScanSaving("my_custom_scansaving")
# create a new dataset ony for the scans in here.
scan_saving.newdataset(None)
scan_saving.dataset.add_techniques("FLUO")
# just prepare a custom scan ...
ls = loopscan(3, .1, mca1, run=False)
s = Scan(ls.acq_chain, scan_saving=scan_saving)
# add some metadata before the scan runs
scan_saving.dataset["FLUO_i0"] = diode1.raw_read
# run the scan[s]
s.run()
# add some metadata after the scan runs
scan_saving.dataset["FLUO_it"] = diode2.raw_read
# just for the debug print at the end
node = scan_saving.dataset.node
# should this print be obligatory?
scan_saving.dataset.check_metadata_consistency()
# close the dataset
scan_saving.enddataset()
# just for diagostics: print all collected metadata
pprint(node.metadata)
# just see if dataset is marked as closed
print("Is closed: ", node.is_closed)
Get metadata from REDIS¶
from pprint import pprint
from blissdata.data.node import get_session_node
def demo_listener(session_name):
session_node = get_session_node(session_name)
for dataset in session_node.walk(wait=False, include_filter="dataset", exclude_children="dataset"):
if dataset.is_closed:
print(f"dataset '{dataset.db_name}' [CLOSED]. The metadata is:")
else:
print(f"dataset '{dataset.db_name}' [RUNNING]. The current metadata is:")
pprint(dataset.metadata)